Speaker: Martí Sànchez-Fibla (Universitat Pompeu Fabra, Barcelona)
Date: 27/04/2023
Time: 10:00
If you would like to attend the seminar, please register here
In this talk, we explore the emergence of collaboration in multiagent reinforcement learning (MARL) through various examples, highlighting the role of human biases such as loss aversion and their impact on collaborative dynamics. We then delve into a paper co-authored by Ricard Sole and Clement Moulin-Frier, which focuses on modeling adaptive dynamics in complex ecosystems. Building on the Forest-Fire model, the study uses fire as an external, fluctuating force in a simulated environment. Agents must balance tree harvesting and fire avoidance, resulting in the evolution of an ecological engineering strategy that optimally maintains biomass while suppressing large fires. We will discuss the implications of these findings for AI management of complex ecosystems, emphasizing the potential benefits of incorporating MARL and collaboration strategies into environmental management and conservation efforts.
Speaker: Madan Rao (NCBS, Bangalore, India)
Date: 20/04/2023
Time: 10:30
If you would like to attend the seminar, please registe here
Speaker: Mukund Thattai (NCBS, Bangalore, India)
Date: 20/04/2023
Time: 10:00
If you would like to attend the seminar, please registe here.
Speaker: Marta Ibañes (Universitat de Barcelona)
Date: 13/04/2023
Time: 10:00
If you would like to attend the seminar, please register here
Along the development of an organism, precursors cells within tissues become specified into distinct cell types. Research in the last decades has shown that many of such processes involve communication between cells, albeit others happen to be mainly cell-autonomous and random. Communication can happen through molecules which move across the tissue or by juxtacrine signaling. Here I will focus on patterning involving diffusion, presenting several theoretical approaches whose predictions are quantitatively compared to data in model organisms. First I will present a mechanism to drive self-confined expression through a diffusing activator. Afterwards I will introduce a new mechanism for diffusion to drive patterning in reaction-diffusion systems.
Speaker: Alejandro Rodriguez-Fraticelli (Institute for Research in Biomedicine, IRB)
Date: 23/03/2023
Time: 10:00
The biologist toolbox has expanded over the last century. It is now common to describe a phenotype using an arsenal of single-cell sequencing and mass spectrometry-based technologies as well as advanced imaging methods. More recently, the merging of single-cell genomics and lineage-tracing innovations has allowed researchers to access cellular ancestry information at unprecedented scale. Applied across systems and scales, cellular phylogenetics is revealing novel insights on developmental dynamics, disease origins and therapeutic responses. This is likely just the tip of the iceberg. As with previous methodological breakthroughs, cellular phylogenetics may hold the potential to revolutionize entire fields and help us engineer enhanced cell therapies. Throughout this talk we will discuss the breadth of findings so far and, through examples of our own, we will cover what we have learned about the biology of blood development, regeneration, aging and cancer.
Speaker: Nuria Lopez-Bigas (Institute for Research in Biomedicine, IRB)
Date: 23/03/2023
Time: 10:00
Our cells accumulate somatic mutations during our lifetime due to multiple mutational processes. Each of these processes leave a specific mutational footprint that can be traced in the form of mutational signatures. Some chemotherapies, for instance, are mutagens and by analyzing whole-genomes from metastases of treated patients we can identify mutational signatures of cancer treatments.
Recent sequencing studies have uncovered abundant clonal expansions driven by somatic mutations in normal human tissues, suggesting that even strong cancer driver mutations are not sufficient for cancer formation. I will introduce the concept of promotion as the rate-limiting step in tumour development and I will explain how we are studying tumor promotion in normal tissues.
Somatic mutations can be used to track the developmental history of cells within an organism. I will explain how we are using somatic mutations for lineage tracing to understand pediatric cancer cases.
During my presentation I will talk about how we use somatic mutations to identify mutational signatures, time clonal expansions, study tumor promotion and perform lineage tracing.
Speaker: James Sharpe (Head of EMBL Barcelona)
Date: 16/03/2023
Time: 10AM CET
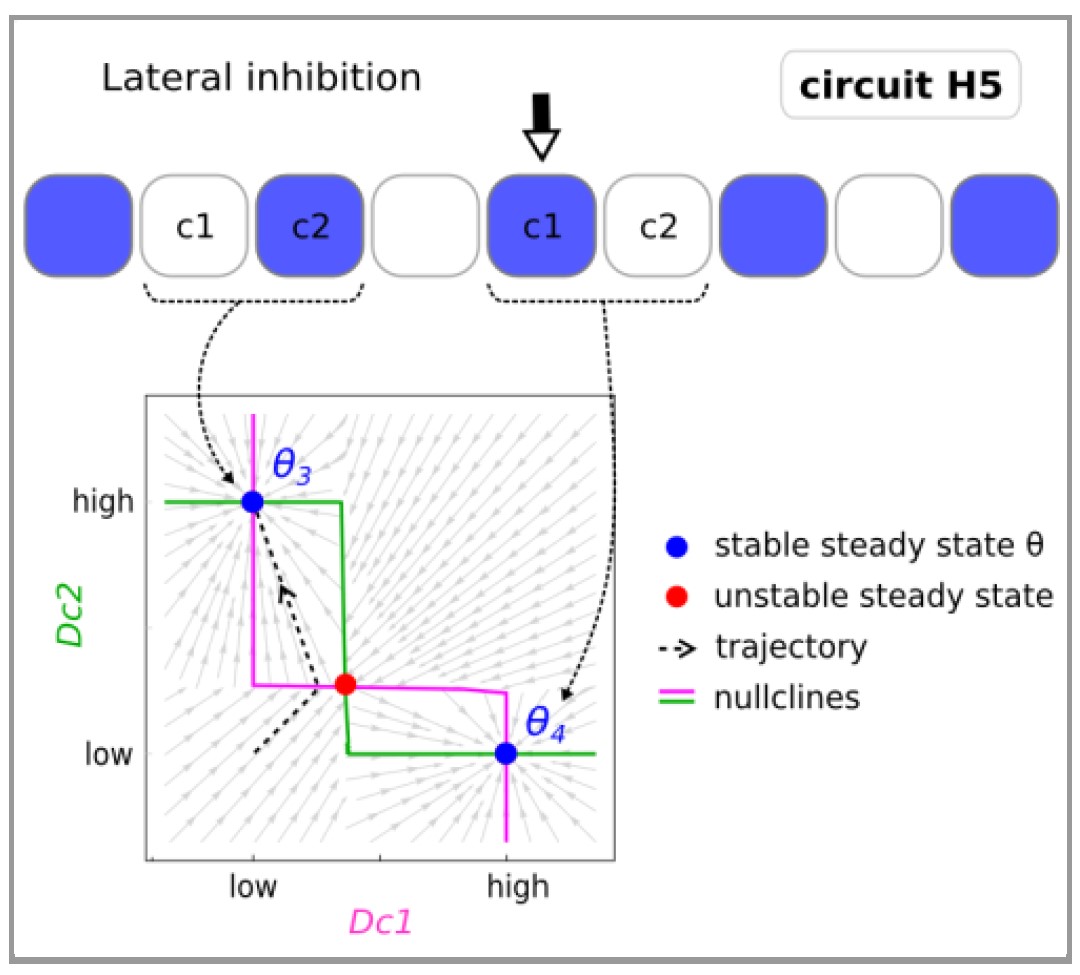
A major challenge in systems biology is to understand the relationship between a circuit's structure and its function, but how is this relationship affected if the circuit must perform multiple distinct functions within the same organism? In particular, to what extent do multi-functional circuits contain modules which reflect the different functions?
Here, we computationally survey a range of bifunctional circuits which show no simple structural modularity: They can switch between two qualitatively distinct functions, while both functions depend on all genes of the circuit.
Our analysis reveals two distinct classes: hybrid circuits which overlay two simpler mono-functional sub-circuits within their circuitry, and emergent circuits, which do not. In this second class, the bi-functionality emerges from more complex designs which are not fully decomposable into distinct modules and are consequently less intuitive to predict or understand. These non-intuitive emergent circuits are just as robust as their hybrid counterparts, and we therefore suggest that the common bias toward studying modular systems may hinder our understanding of real biological circuits.
Speaker: Gonzalo Parra (Barcelona Supercomputing Centre)
Date: 09/03/2023
Time: 10AM CET
Energetic local frustration offers a biophysical perspective to interpret the effects of sequence variability on protein families. Here we present a novel methodology [1] to analyze local frustration patterns within protein families that allows us to uncover constraints related to stability and function, and identify differential frustration patterns in families with a common ancestry. We have analyzed these signals in very well studied cases such as PDZ, SH3, alpha and beta globins and RAS families. Recent advances in protein structure prediction make it possible to analyze a vast majority of the protein space. An automatic and unsupervised proteome-wide analysis on the SARS-CoV-2 virus demonstrates the potential of our approach to enhance our understanding of the natural phenotypic diversity of protein families beyond single protein instances. We have applied our method to modify biophysical properties of natural proteins based on their family properties, as well as perform unsupervised analysis of large datasets to shed light on the physicochemical signatures of poorly characterized proteins such as emergent pathogens.
[1] Freiberger, M. I. et al. The Evolution of Local Energetic Frustration in Protein Families. BioRXiv 2023 at https://doi.org/10.1101/2023.01.25.525527__;!!D9dNQwwGXtA!UE3kotrrTKNiWH550N-pa-6Kc8waucJ-C3juoSP25bHG02BeQOPcfosnHlpTldII0bj3xUY72VAuJjSm_ph4nv66$
Speaker: Alejandro Torres-Sanchez (EMBL Barcelona)
Date: 02/03/2023
Time: 10AM CET
I will discuss Onsager’s variational principle, an extension by Onsager of the principle of least energy dissipation by lord Rayleigh. Resorting to examples in biology, I will show that this variational approach is a very effective formalism to describe linear and non-linear non-equilibrium processes combining multiple physical phenomena. In particular, I will show how this principle can be employed to derive the governing equations that regulate the fracking of cell adhesions by osmotic pressure. Finally, I will show that this principle provides a direct route for the time and space discretisation of the equations needed for numerical simulations.



